BACKGROUND
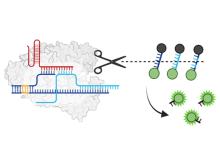
CRISPR-based diagnostics have rapidly emerged as powerful tools for nucleic acid detection due to their programmability, sensitivity, and adaptability to point-of-care settings. Central to these systems is the phenomenon of collateral cleavage, where CRISPR-associated enzymes such as Cas12 and Cas13, once activated by a specific target sequence, indiscriminately cleave nearby single-stranded nucleic acids. This mechanism underpins diagnostic platforms like DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reporter), HOLMES (One-Hour Low-cost Multipurpose Highly Efficient System), and SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing), which utilize fluorescent or lateral flow reporters to signal the presence of target DNA or RNA.
TECHNOLOGY
Researchers at the University of Toronto have developed a novel class of chimeric reporters that significantly improve the RNA trans-cleavage efficiency of Cas12 enzymes, offering a robust alternative to traditional ssDNA reporters. The chimeric reporters are composed of RNA bases interspaced with modified DNA sugars and bases. The system is compatible with multiple Cas12 variants (LbCas12a, AsCas12a, AapCas12b) and supports both amplification-assisted and amplification-free detection methods. It also accommodates diverse reporting mechanisms, including fluorescence, lateral flow assays, and colorimetric outputs. Experimental results demonstrate that chimeric reporters achieve similar or superior performance compared to ssDNA reporters, with enhanced signal-to-noise ratios.
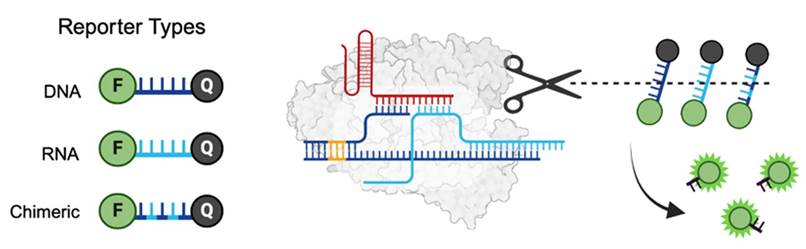
Figure 1. Collateral cleavage using DNA, RNA, and Chimeric reporters. Once activated by a target sequence, CRISPR-associated enzymes indiscriminately cleave nearby single-stranded nucleic acids releasing an optical reporter.
COMPETITIVE ADVANTAGE
- Enhanced RNA Cleavage: Chimeric sequences significantly improve RNA trans-cleavage kinetics, overcoming the inherent RNase limitations of Cas12 proteins.
- Comparable or Superior to ssDNA: Achieves similar or better cleavage kinetics and limits of detection (LOD) compared to ssDNA reporters.
- High Signal-to-Noise Ratio: Especially with polyrUA chimeric sequences, enhancing detection clarity and reliability.
- Broad Cas12 Compatibility: Effective across multiple Cas12 variants (LbCas12a, AsCas12a, AapCas12b), with optimized performance for each.
APPLICATIONS
- Point-of-care diagnostics for infectious diseases (e.g., HPV, SARS-CoV-2)
- Environmental and agricultural pathogen surveillance
INTELLECTUAL PROPERTY STATUS
- PCT Application (Feb 2025)
PROJECT STATUS
The technology has been validated in vitro using multiple Cas12 proteins and synthetic targets. Experimental data confirm enhanced cleavage kinetics and detection sensitivity. The system has been demonstrated in both amplification-free and amplification-assisted formats, including micro-scale droplet visualization.
KEYWORDS
CRISPR diagnostics, Cas12a, Cas12b, chimeric nucleic acids, RNA detection, ssDNA, synthetic biology, point-of-care, lateral flow assay, nucleic acid biosensor, programmable diagnostics, pathogen, virus, bacteria