BACKGROUND
Traditional computational methods face challenges in detecting ligandable cryptic and surface sites in the "undruggable" proteome. Chemoproteomics, using covalent probes, offers promise for targeted therapeutic intervention of the “undruggable” proteome and overcoming resistance mechanisms. However, current methods primarily focus on cysteine residues, representing only about 20% of the quantifiable proteome. There is a need for advanced computational tools to map ligandability beyond cysteine residues, such as at lysines and tyrosines.
TECHNOLOGY
Researchers at the University of Toronto have developed a ligandability assessment tool for covalent inhibitor development. It involves a computational platform that validates empirically profiled chemoproteomics sites and facilitates target ligandability nomination (Figure 1). This platform uses conformational sampling to detect hidden or unformed ligandable sites to and perform tractability assessment on covalent binding sites, enhancing the accuracy of in-vitro ligandability validation trials. The platform has been validated for cysteine-, lysine-, and tyrosine-based chemoproteomics sites for both apparent and cryptic ligandable cavities.
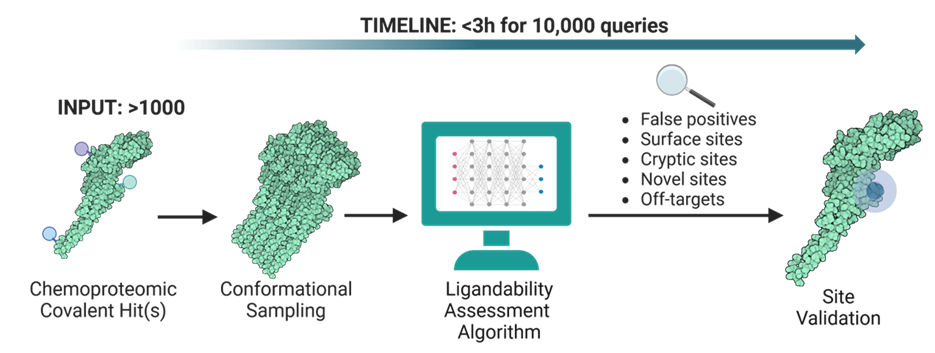
Figure 1. Covalent Hot Spot Assessment. Chemoproteomic sites are used as inputs, conformational sampling in conducted, each conformer is passed thorough the Ligandability Assessment Algorithm, and possible ligandable sides are determined.
COMPETITIVE ADVANTAGE
- Residue-Agnostic: Capable of assessing ligandability surrounding any site including cysteine, lysine, and tyrosine residues.
- High Accuracy: Demonstrated 98% accuracy in identifying covalent ligandable sites on holo protein structures and 90% on apo protein structures
- Rapid Assessment: Processes chemoproteomics data at a rate of 1 query per second.
- Comprehensive: Integrates omics enrichment for target prioritization and validation.
- Cost-Effective: Reduces the need for extensive wet-lab validation, saving time and resources.
APPLICATIONS
- Drug Discovery: Identifying and validating novel therapeutic targets in the "undruggable" proteome.
- Target Prioritization: Facilitating rational design of targeted covalent inhibitors (TCIs).
- Omics Analysis: Providing insights into the functional and disease-linked impact of targeting specific sites.
INTELLECTUAL PROPERTY STATUS
- Trade Secret and Know-How
PROJECT STATUS
The platform has been validated using established holo, apo, and cryptic binding sites on various protein structures. It has successfully assessed and mass-validated cysteine, lysine, and tyrosine-based chemoproteomics sites for ligandable cavities. These include an active site on TGM2 and a covalent site on PLK1.
KEYWORDS
Chemoproteomics, targeted covalent therapeutics, ligandability mapping, dark proteome, computational drug discovery, machine learning.