BACKGROUND
Gene therapy using space-constrained delivery vehicles like adeno-associated viral (AAV) vectors can benefit from the shortening of large tissue-specific promoters. This motivated efforts to make promoters highly compact while maintaining activity and specificity. However, the promoter design process is largely iterative and experimental. It typically involves the synthesis of a promoter fragment roughly up to 3000 nucleotides upstream and ∼50 nucleotides downstream of the transcription start site. Upstream fragments are then truncated until loss of function occurs, indicating the optimal short promoter. However, it may still contain “subsequences” that do not contribute to function. If an alternative solution can be designed to computationally predict the transcription factor binding sites, this would enable removal of non-functional “subsequences” yielding even shorter promoters and in a cost-effective manner.
TECHNOLOGY
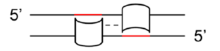
Researchers at the University of Toronto have developed an algorithm to design synthetic short promoters by concatenating predicted transcription factor binding motifs. The algorithm works by identifying probable palindromic sequences, that transcription factors generally bind to, through utilizing an individual nucleotide scoring system. A high density of probable palindromes (Figure 1) is usually correlated to the key elements of the enhancer or repressor regions responsible for promoter function. Concatenation of these regions upstream of the transcription start site to a core promoter is used to generate the synthetic short promoters. Using this algorithm, greater than 20 model short promoters have been tested in cells and a library of putative synthetic short promoters has been generated for all promoters in the human and mouse genomes.
Figure 1. Binding of direct and indirect oligomeric transcription factors to a promoter sequence. Red lines indicate palindromic sequences, while dotted lines indicate protein-to-protein interactions.
COMPETITIVE ADVANTAGE
- Facile method to construct short promoters
- General reduction in promoter length by ~66%
- A collection of constitutive, inducible, tissue-specific and cell-specific promoters were tested without loss of function (e.g. CMV, mouse synapsin-1, human EF1)
- Short promoters predicted for the entire human and mouse genomes
APPLICATIONS
- Short promoters for gene therapy
INTELLECTUAL PROPERTY
- Provisional patent application filed (Feb 2022)
PROJECT STATUS
A computation method has been designed to identify potential transcription factor binding sites to generate short promoters. It has been experimentally validated in cell lines (HEK293 and N2A) for activity and specificity by using the CMV and the mouse synapsin-1 promoters (Figure 2). Functionality has been further verified with 11 other short promoters.
Figure 2. Comparative activity of full-length and short promoters. Full-length promoters (CMVp, mSyn1p) showed statistically identical expression levels to the short promoters (PCVMp, PmSyn1p) indicating no loss in functionality. CMV experiments were conducted in HEK293 cells, while Syn experiments were conducted in a neuronal cell line (N2A).