BACKGROUND
Protein-protein interactions (PPIs) govern many cellular processes and play a crucial role in organism function. There are estimated to be between 130,000-600,000 types of PPIs in the human ‘interactome’. Aberrant interactions have been linked to various diseases like cancer, infectious and neurodegenerative diseases. As such, the identification and modulation of PPIs is an attractive approach to the development of new drugs. There are several techniques available (LuTHy, LUMIER, MAPPIT, KISS, Yeast Two-Hybrid (YTH), PCA, and NAPPA) for the identification of PPIs and these can often be also utilized for drug screening. However, all of them exhibit certain biases that prevent coverage across the human ‘interactome’, thus necessitating the development of complementary techniques.
TECHNOLOGY
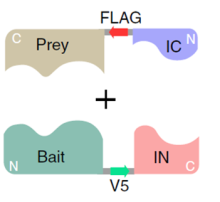
Researchers at the University of Toronto have developed a live cell approach called Split Intein-Mediated Protein Ligation (SIMPL) to extend PPI detection coverage (Figure 1). In this method, bait and prey proteins are fused to one of two fragments composing an intein. Interaction of the bait and prey leads to reconstitution of the split intein, instigation a splicing event that ligates the bait to the prey. This larger sized fragment can then be easily detected using Western Blot analysis. Alternatively, slight modifications to the approach, namely the incorporation of tags (FLAG, V5), can enable a high throughput ELISA-based screening method.
Figure 1. The Split Intein-Mediated Protein Ligation (SIMPL) process. IC and IN indicted the C and N-terminal of the split intein (I). Binding of the bait and prey initiates enzymatic ligation, joining the bait and prey and excising the split intein. Detection of the PPI event can be achieved using a Western blot or ELISA.
COMPETITIVE ADVANTAGE
- High sensitivity and specificity
- Dose-response range similar to BRET (i.e. 1 nm rapamycin concentration induced interaction between FRB and FKBP1A)
- 41-56% of PPIs detected in reference set (88 possible PPIs) with false-positive rate of ~5%
- Robust
- Works in mammalian cells and animal models (e.g. C. elegans)
- Capable of tracking fast PPIs (i.e. ~2 min)
- Capable of detecting transient or weak PPIs
- Resistant to harsh experimental conditions and multiple processing steps (i.e. ligation is irreversible)
- No cofactors or energy source required
- High throughput ELISA-based format available
- Live cell-based assay for drug screening
- Sub-cellularly compartment detection possible
APPLICATIONS
- Protein-protein interaction detection
- Drug screening platform
INTELLECTUAL PROPERTY STATUS
- National Phase: CA, EU, US (June 2020)
PROJECT STATUS
Proof-of-concept studies have been conducted. The SIMPL system has been developed and tested using rapamycin-induced heterodimerization of FKBP1A and the FKBP rapamycin-binding (FRB) domain of mTOR12 in HEK 293 and isogenic stable cell lines. Orientational dependence of fusion of bait/prey to the split intein has been evaluated. SIMPL has been benchmarked using a reference set containing 88 available positive PPIs. Its feasibility has been evaluated in a multicellular animal model (C. elegans) and it has been tested as a drug screening platform.