BACKGROUND
Protein phosphorylation is the most frequent post-translational modification and plays a critical role in regulating signal transduction pathways that are essential for cell growth, migration, and replication. Since aberrant protein phosphorylation has been implicated in numerous diseases including cancer, mapping of and determining changes in the protein phosphorylation profile is important for uncovering pathogenic signalling pathways and identifying novel therapeutic strategies. However, general methods struggle with determining a comprehensive profile of the phosphoproteome due to dynamic range and detection sensitivity issues. These sampling deficiencies are particularly acute for the analysis of the phosphotyrosine (pTyr)-containing proteome, due to its rapid turnover and low cellular abundance. Additionally, pTyr specific methods (i.e. antibodies) are poorly scalable, require optimization of experimental conditions, and suffers from batch-to-batch variations.
TECHNOLOGY
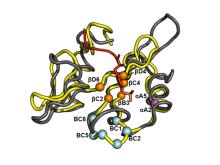
Researchers at the University of Toronto have proposed a set of cost-effective pTyr enrichment tools (AffiniPure) that can recover a greater percentage of the pTyr-proteome. The technology relies on a set of SH2 domains, which bind pTyr residues, that are bioengineered to be superbinders (Figure 1). Several residues in the pTry binding pocket of Src and Fes SH2 domains were chosen for optimization using a phage display library comprising 16 billion unique combinations. The resulting Src and Fes superbinder motifs were then incorporated into 18 other SH2 domains to produce a panel of superbinders with different specificity profiles for probing the pTyr phosphoproteome with unprecedented depth and coverage than is possible with current tools (antibodies or wild-type SH2 domains).
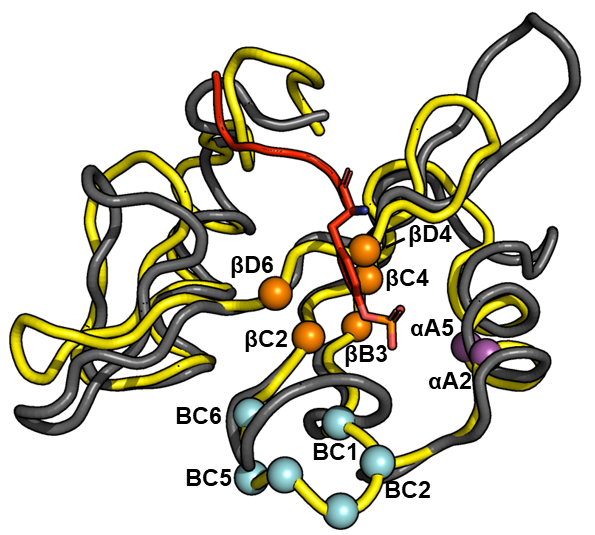
Figure 1. Superposition of the structure of Fes-SH2 (yellow) and Src-SH2 (grey) in complex with a pTyr-containing peptide. Positions that were varied in the phage display screen are shown as spheres colored magenta, orange or cyan. The pTyr side chain of the peptide ligand is shown as a red stick.
COMPETITIVE ADVANTAGE
- Display 500 to 3000-fold enhancement in pTry binding affinity compared to wildtype
- Each superbinder generally isolates 10x to 20x more pTry peptides from cells than corresponding wild-type SH2 domains
- Combinations of the best superbinders can be packaged to enrich unique subsets of phosphotyrosine peptides
APPLICATIONS
- Phosphotyrosine proteome determination
INTELLECTUAL PROPERTY STATUS
- Provisional patent filed (June 2021)
PROJECT STATUS
Proof of concept studies have been conducted. Phage display was used to optimize binding of phosphotyrosine peptides through variation of thirteen residues in the binding pocket of Src and Fes SH2 domains. A panel of 18 additional SH2 domains were selected to receive the optimized Src or Fes superbinder motifs. Pull-down assays were conducted using superbinder-bound magnetic beads in unstimulated and stimulated K562 cell lysate (Figure 2) and further benchmarked to commercially available Zr/Ti IMAC beads.
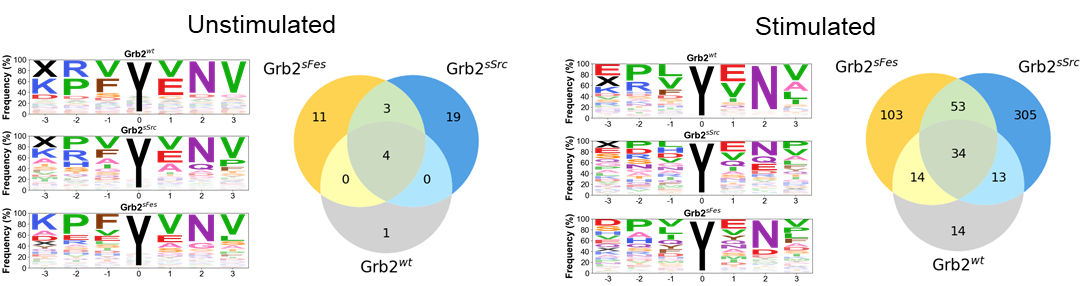
Figure 2. Illustrative example of improvement in isolation of phosphotyrosine peptides by a superbinder. Grb2-SH2 superbinders containing the super-Fes (Grb2sFes) and super-Src (Grb2sSrc) binding motifs are compared to the wild-type Grb-SH2 domain (Grb2wt) in unstimuated and stimulated K562 cell lysates. Venn diagrams show considerably greater (7x to 20x) isolation of unique peptides than wild-type SH2 domains. Peptide selectivity is shown in the associated frequency graphs.