BACKGROUND
Protein sequencing is essential for understanding a protein’s structure, function, and interactions within biological systems. Traditional methods, such as Edman degradation and mass spectrometry, have been instrumental in advancing the field, providing insights into protein function, disease mechanisms, and therapeutic development. However, there remains a need for novel methods and assays capable of sequencing single polypeptide molecules and identifying, quantifying, and imaging many different proteins simultaneously in complex biological samples. Addressing these challenges is critical for advancing proteomics, especially in the context of systems biology, where comprehensive and precise protein analysis is increasingly vital.
TECHNOLOGY
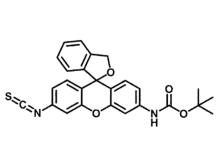
This technology provides molecular imaging–based methods, assays, and reagents that are useful for sequencing proteins. An optically active probe (HMRG-BOC-ITC; Figure 1) is first conjugated to the N-terminal amino acid of the polypeptide. The probe exhibits small but characteristic differences in spectral properties when conjugated to different amino acids, thereby allowing identification of all 20 amino acids. Sequencing can be achieved through a cycle of repeated cleavage of the N-terminal amino acid followed by conjugation of the newly exposed N-terminal amino acid to the probe. Furthermore, the use of single-molecule imaging techniques (e.g., super-resolution microscopy), which allow for the detection of spectral properties from spatially resolved individual molecules, may be used for the parallel sequencing of large numbers of polypeptides in vitro or in situ, such as within tissues, cells, lipid membranes, or organelles. This enables information to be obtained on protein identity, quantity, and subcellular location within a biological or environmental sample.
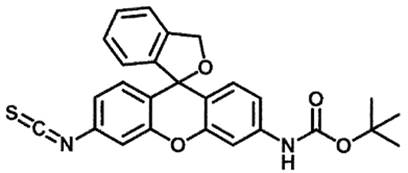
Figure 1. Chemical structure of the probe hydroxymethyl rhodamine green (HMRG) with a reactive isothiocyanate (ITC) group and a tert-Butyloxycarbonyl (BOC) protecting group.
COMPETITIVE ADVANTAGE
- High Throughput: Enables rapid simultaneous sequencing of large numbers (hundreds of thousands) of proteins, enhancing efficiency and scalability.
- In Situ Sequencing: Protein sequencing can be conducted on a slide or flow cell, at the plasma membrane, or within intracellular organelles of a cell, thereby revealing the composition, localization and physical interactions of proteins that are present.
- Optimized Reagents: Utilizes specialized chemical reagents that facilitate efficient cleavage and identification of amino acids.
APPLICATIONS
- Proteomics Sequencing: Provides accurate and efficient protein sequencing for studying protein structure and function.
INTELLECTUAL PROPERTY STATUS
- National Phase Applications: CA, EP, US (US11268963B2)
PROJECT STATUS
Proof-of-concept studies have been conducted. Studies have been conducted on the surface of borosilicate coverglass and using peptide beads.
KEYWORDS
Protein sequencing, amino acid identification, high-throughput sequencing, proteomics, molecular biology, chemical reagents.