BACKGROUND
Determining and modifying protein-protein interactions (PPIs) are important because their alterations are involved in various diseases. Although numerous techniques are available for studying PPIs, each one is accompanied by various limitations (e.g. high cost, requirement for labeling, inability to recapitulate in vivo conditions, incompatibility with HTS formats and the requirement for highly specialized equipment and expertise). To maximize the capability to explore PPIs, our researchers recently invented Split-Intein Mediated Protein Ligation (SIMPL) where a PPI association event is coupled to intein-mediated ligation. However, the ELISA readout conventionally used with this assay is expensive, time-consuming and cumbersome necessitating an improved detection system.
TECHNOLOGY
Researchers at the University of Toronto have developed an improved screening system for detection of protein-protein interactions (PPIs) using Split-Intein Mediated Protein Ligation and tri-part NanoLuc (SIMPL-tNLuc or SIMPL2) (Figure 1). In this system, the β10 tag from tNLuc is fused to the bait protein and the N-terminal of a GP41-1 split intein, while the β9 tag is fused to the prey protein and the C-terminal of the split intein. Interaction of the bait (B) and prey (P) proteins induces splicing by the reconstituted intein, resulting in ligation of B and P. This simultaneously also places the β10 and β9 tag in tandem with each other. Addition of the third fragment, Δ11S, needed to reconstitute NanoLuc results is a functional enzyme capable of producing a luminescent readout indicative of the PPI event.
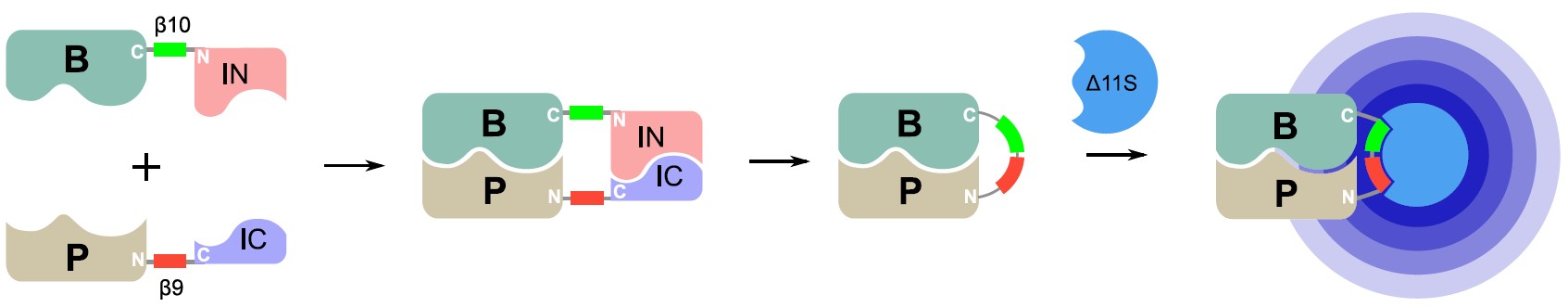
Figure 1. Detection of protein-protein interactions (PPIs) using SIMPL2. Interaction of the bait (B) and prey (P) proteins results in a ligation event mediated by the split intein that places two fragments of NanoLuc (β10 and β9) in close proximity to each other. Addition of the third Δ11S fragment reconstitutes the NanoLuc protein whose restored luminescent functionality is indicative of a PPI.
COMPETITIVE ADVANTAGE
- Improved high-throughput screening platform for detecting PPIs and their inhibitors
- Cheaper and less cumbersome that SIMPL-ELISA
- Eliminates the need for the numerous cycles of aspiration and washing (i.e. allows for detection in the homogeneous liquid phase)
- Expensive reagents such as labelled antibodies not required
- Improved quantifiability compared to SIMPL-ELISA
- One order of magnitude improved sensitivity
- Reduced non-specific splicing
- 5-6-fold improvement in signal/background ratio
- One order of magnitude improved sensitivity
- More sensitive in detecting weak PPIs or induced proximity interactions compared to NanoBiT assay
- Cheaper and less cumbersome that SIMPL-ELISA
APPLICATIONS
- HTS of protein-protein interactions
- Drug Discovery
- Inhibitors of PPIs
- Molecular glues
- ProTACs
INTELLECTUAL PROPERTY STATUS
- US provisional (Apr 2024)
PROJECT STATUS
Proof-of-concept studies have been conducted to demonstrate the ability of SIMPL2 to detect PPIs and act as a drug discovery tool. SIMPL2 has been evaluated using a reference PPI set (hsPRS-v2) where 48% of interactions were clearly identified at a threshold above background. The ability to detect small molecule modulators of PPIs has been demonstrated using covalent (sotorasib, afatinib) and non-covalent (AG1478, BI-2865) inhibitors, a molecular glue (RO-5963) and a PROTAC (ARV-825).