BACKGROUND
Serum-free cell culture media design is important because it offers the opportunity to construct chemically defined media that possess greater batch-to-batch reproducibility and clinically compatible reagents; benefits not necessarily associated with serum-containing media. As such, it is an ideal method for producing media for the growing cell therapy market segment where high quality and reproducibility standards influence clinical translation. However, the use of defined chemical factors often creates a complex, large-scale optimization problem where the number of possible combinations of factors across different dose concentrations creates a search space so large that only a millionth or less of it can be sampled. Consequently, this renders traditional media optimization techniques ineffective and necessitates the development of new techniques to handle this problem.
TECHNOLOGY
Researchers at the University of Toronto had previously constructed a high dimensional-differential evolution (HDDE) algorithm to optimize media containing as many as 15 different chemical factors. They used it to produce serum-free media for the expansion of both TF-1 and T cells that is comparable to gold standard media. More recently, they have added an artificial neural network module to the HDDE algorithm (HiDiNeu) enabling both faster determination of optimal media composition and optimization of exponentially larger search spaces (Figure 1). HiDiNeu has been tested in silico using standard benchmark functions where it was shown to reach a close-to-optimum formulation in only two generations (2x quicker than the HDDE algorithm) and with higher fidelity to the true solution. Use of HiDiNeu also enabled superior optimization of T cell expansion than HDDE. It’s also important to note that such algorithms can optimize any measurable cell parameter beyond cell growth and maintenance (e.g. contractility, differentiation potential, etc.).
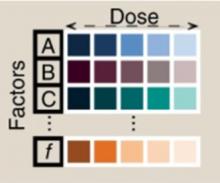
Figure 1. Serum-free cell media optimization using algorithms. For a given number of factors (A, B, C, . . f) and dose concentrations (coloured squares), a small subset of factor combinations (compared to the total search space) is generated for experimental testing. From these experimental results, a model of the search space is generated using an automated AI algorithm (i.e. requiring no user input) to assess optimization. A decision is then taken to either suggest further experiments to more accurate define the search space or, if an optimal media constitution is identified, the loop is halted with the optimized media solution proposed.
COMPETITIVE ADVANTAGE
- Serum-media optimization achieved with miniscule sampling of the solution space
- <5x10-6 % of the total search space
- Optimization of large number of factors possible
- ~ 25 chemical factors or greater
- AI module increases speed, reduces work, and increases accuracy of solution
- 2x faster
- 2x less experimental work needed
- >50% reduction in error of solution (compared to non-AI algorithms)
- Highly robust to data variance
APPLICATIONS
- Serum-free media design and optimization
- Cell manufacturing processes
INTELLECTUAL PROPERTY STATUS
- Provisional patent application filed (Dec 2023)
PROJECT STATUS
HiDiNeu has been tested in silico on three high-dimensional benchmark functions (Rosenbrock, Ackley, Rastrigen) and has also been used to generate serum-free media for T cell expansion that is comparable to gold standard serum-based media (Figure 2).
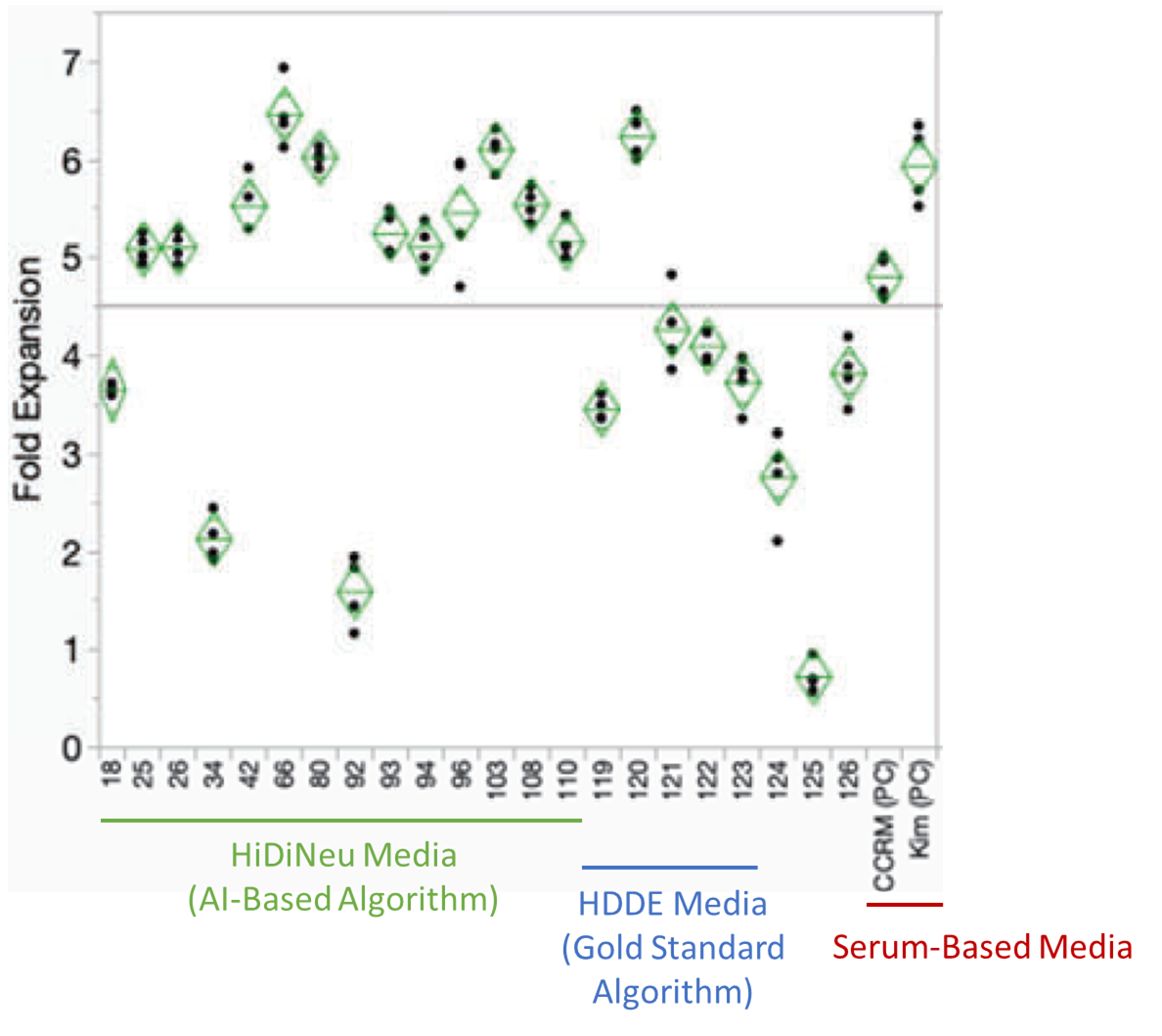
Figure 2. T cell expansion levels from several top formulations generated using HiDiNeu, HDDE (gold standard algorithm), and Serum-Based controls. HiDiNeu was able to generate several formulations of comparative expansion potential to serum-based controls.