BACKGROUND
Protein sequencing is essential for understanding biological processes, disease mechanisms, and therapeutic development. Traditional protein sequencing methods, such as Edman degradation and mass spectrometry, have been instrumental in advancing proteomics. However, these techniques come with significant limitations. Edman degradation is labor-intensive and inefficient for long or modified proteins, while mass spectrometry, though powerful, often struggles with complex mixtures, low-abundance proteins, and post-translational modifications. Moreover, both methods typically require bulk samples, making them unsuitable for analyzing rare proteins or single-cell proteomes. As the field of biology moves toward more personalized and precision-based approaches, there is a growing demand for technologies that can sequence proteins at the single-molecule level and for simultaneous parallel sequencing of large-numbers of polypeptides.
TECHNOLOGY
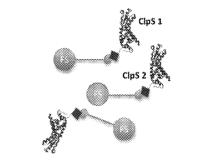
Researchers at the University of Toronto have introduced a novel method for sequencing polypeptides at the single-molecule level. This approach involves the use of a set of optical probes comprising variants of CIpS proteins from E. coli each capable of selectively binding a specific N-terminal amino acid. Conjugation of these proteins to a unique optical readout (e.g. FluoSpheres) allows the determination of the specific amino acid identity. Cleavage of the N-terminal residue and repetition of the process enables the determination of the sequence of an immobilized polypeptide. Parallel detection of multiple polypeptides can be achieved by affixing them to a surface (e.g. glass slide) in a spatially resolved manner.
COMPETITIVE ADVANTAGE
- Single-Molecule Sensitivity: Enables sequencing of individual protein molecules, improving resolution and reducing sample requirements.
- Residue-Specific Probes: Uses a library of probes with high specificity for specific N-terminal amino acids or derivatives.
- Scalable and Parallelizable: Suitable for high-throughput applications and multiplexed analysis.
- Robust Sample Tolerance: Cell extract or tissue extract, biological fluid (e.g. serum, plasma), soil, single cell, etc.
APPLICATIONS
-
Proteomics Research: Provides accurate and efficient protein sequencing for studying protein structure and function.
INTELLECTUAL PROPERTY STATUS
- Granted US Patent (US10481162B2)
PROJECT STATUS
Proof-of-concept studies have been conducted.
KEYWORDS
Protein sequencing, single-molecule analysis, N-terminal probes, proteomics, biomarker discovery, biopharmaceuticals.